On this page... (hide)
- 1. Current Projects
- 1.1 EPRSC. “Learning MRI and histology image mappings for cancer diagnosis and prognosis”. 15/12/2017 – 14/12/2020.
- 1.2 EPRSC Fellowship. “Non-invasive MRI biomarkers for Oncology”. 1/07/2016 – 30/06/2021.
- 1.3 EPRSC. “A biophysical simulation framework for magnetic resonance microstructure imaging”. 1/04/2016 – 31/03/2019.
- 1.4 EPSRC Doctoral Prize Fellowship. “Quantitative diffusion MRI for prostate cancer grading”. 1/01/2016 – 01/01/2018.
- 1.5 Prostate Cancer UK. “CombIning advaNces in imagiNg with biOmarkers for improVed diagnosis of Aggressive prosTate cancEr - INNOVATE”. 1/01/2016 - 31/12/2017.
- 1.6 Microsoft Research PhD Studentship. “Image Quality Transfer”. 1/10/2015 – 28/09/2019.
- 1.7 NIH. “Structure and Function of the Placenta from Implantation to Delivery. A Next Generation MRI Approach”. 17/09/2015 - 31/08/2018.
- 1.8 EU Horizon 2020. “A Clinical Decision Support system based on Quantitative multimodal brain MRI for personalized treatment in neurological and psychiatric disorders (CDS-QUAMRI)”. 1/09/2015 – 01/09/2020.
- 1.9 Magnetic Resonance Imaging in Multiple Sclerosis (MAGNIMS). “To explore imaging signature of multiple sclerosis phenotypes”. 1/07/2015 – 01/07/2016.
- 1.10 EPRSC. “National Facility for In Vivo MR Imaging of Human Tissue Microstructure”. 1/06/2015 – 31/05/2020.
- 1.11 EPSRC. “Anatomy driven brain connectivity mapping”. 1/06/14 – 31/05/17.
- 1.12 MRC. “In vivo microstructural neuroimaging in infants at risk of developing neurocognitive delay or neurobehavioural disorders”. 1/04/14 – 31/03/19.
- 1.13 UCL CS Research Excellence Studentship. "Automatic analysis of histology with machine learning". 10/03/2014 — 01/09/2017.
- 1.14 UCL SLMS Grand Challenge Studentships project 2013. “Parcellation of the human cortex using quantitative MRI”. 1/10/2013—30/09/2016.
- 1.15 EPSRC Breast cancer project. “Medical imaging markers of cancer initiation, progression and therapeutic response in the breast based on tissue microstructure”. 1/01/13 – 31/12/15.
- 1.16 UCL SLMS Grand Challenge Studentship project. "Development and validation of machine learning techniques to facilitate diagnosis and predict prognosis in patients with Multiple Sclerosis". 16/11/12-15/11/15
- 2. Completed Projects
- 2.1 CMIC Platform Grant (pump-priming Award). “Enabling multi-site high precision spinal cord MRI”. 01/06/2017 – 01/06/2018.
- 2.2 UCL SLMS Grand Challenge Studentships project 2012. “Axonal density as MR imaging biomarker: from bench to bedside”. 1/10/2012—30/09/2015.
- 2.3 EPSRC Brain connectivity project. “Robust graph analysis of brain connectivity”. 1/06/2012—31/05/2015.
- 2.4 Leverhulme Trust Fellowship. "New imaging techniques to reveal the relationship between brain tissue microstructure and brain function". 1/05/2012-31/12/2015.
- 2.5 EPSRC Healthcare Partnerships Project. "Axon and myelin damage assessed using advanced diffusion imaging: from mathematical models to clinical applications". 1/09/2011-31/08/2014.
- 2.6 EPSRC Program grant. “Intelligent Imaging: Motion, Form and Function Across Scale”. 1/06/2010—31/05/2015.
- 2.7 EPSRC Leadership Fellowship. "Direct measurements of microstructure from MRI." EP/E007748. 1/10/2008—30/09/2014.
- 2.8 European Commission (Future and Emerging Technologies program). “CONNECT: Consortium of NeuroImagers for the Non-invasive Exploration of Brain Connectivity and Tractography”. 1/01/2010—31/10/2012.
- 2.9 International Spinal Research Trust (ISRT). "Spinal Cord Diffusion Imaging: Challenging Characterization and Prognostic Value'' Natalie Rose-Barr Studentship. 1/10/2008--30/9/2011.
- 2.10 EPSRC. "Monte Carlo random effects modelling in diffusion MRI: a new window on microstructure and white matter architecture.'' EP/G025452/01. 1/10/2008--30/9/2011.
- 2.11 EPSRC CASE Studentship with GSK. 28/9/2008--27/9/2011.
- 2.12 EPSRC Doctoral Prize (PhD+). “Differentiating grades of brain tumour with DW-MRI”. 1/01/2011-1/01/2012.
- 2.13 EPSRC. "Monte-Carlo simulation framework for diffusion MRI.'' EP/E056938/1. 1/9/07--31/8/10.
- 2.14 NHS, Frenchay Hospital. "The optimisation of diffusion imaging to quantify fluid flow within the central nervous system'' 1/1/2008--31/12/2009.
- 2.15 EPSRC CASE Studentship. 1/10/2005--31/9/2008.
- 2.16 EPSRC. "Multiple-fibre reconstruction in diffusion MRI''. GR/T22858/01. 1/1/2005--31/12/2007.
- 2.17 EPSRC EngD studentship (Sortex). 1/11/2004--31/10/2007.
- 2.18 EPSRC EngD studentship (Philips Medical Systems). 1/10/04--30/9/07.
- 2.19 EPSRC Fast-stream grant. "Registration of DT MRI''. GR/R13715/01. 1/2/2001--31/1/2004.
- 2.20 EPSRC/DTI E-science intiative. 1/10/2001--31/3/2002.
1. Current Projects
1.1 EPRSC. “Learning MRI and histology image mappings for cancer diagnosis and prognosis”. 15/12/2017 – 14/12/2020.
- Personnel: Francesco Grussu, Thomy Mertzanidou, Eleftheria Panagiotaki, Daniel Alexander.
- Summary: This project aims to exploit recent advances in machine learning to address acute problems in cancer management – most directly prostate cancer. We will obtain data relating i) MRI and histology images, and ii) histology and patient outcome. In combination, these support a two-step learning and estimation process: from MRI to histological features; and from histological features to patient prognosis. Such mappings can provide invaluable new information for clinical decision making, as well as guide the design of maximally informative future MRI protocols. The summary of the project is on the EPSRC website here.
1.2 EPRSC Fellowship. “Non-invasive MRI biomarkers for Oncology”. 1/07/2016 – 30/06/2021.
- Personnel: Eleftheria Panagiotaki
- Summary: This project develops innovative Magnetic Resonance Imaging (MRI) methods to reveal new non-invasive markers of cancer pathology. In particular the research programme develops biophysical models of tumour tissue that support non-invasive estimates of key characteristics of tissue cellular architecture including those that currently guide diagnosis, grading, and treatment assignment through classical histology. The summary of the project is on the EPSRC website here.
1.3 EPRSC. “A biophysical simulation framework for magnetic resonance microstructure imaging”. 1/04/2016 – 31/03/2019.
- Personnel: Gary Zhang, Simon Walker-Samuel, Karin Shmueli, Daniel Alexander.
- Summary: This project extends the diffusion simulation in Camino to include a range of new effects including flow, permeability, and susceptibility. It makes the first steps towards developing multi-modal MR tissue models and next-generation imaging techniques that exploit them. The summary of the project is on the EPSRC website here.
1.4 EPSRC Doctoral Prize Fellowship. “Quantitative diffusion MRI for prostate cancer grading”. 1/01/2016 – 01/01/2018.
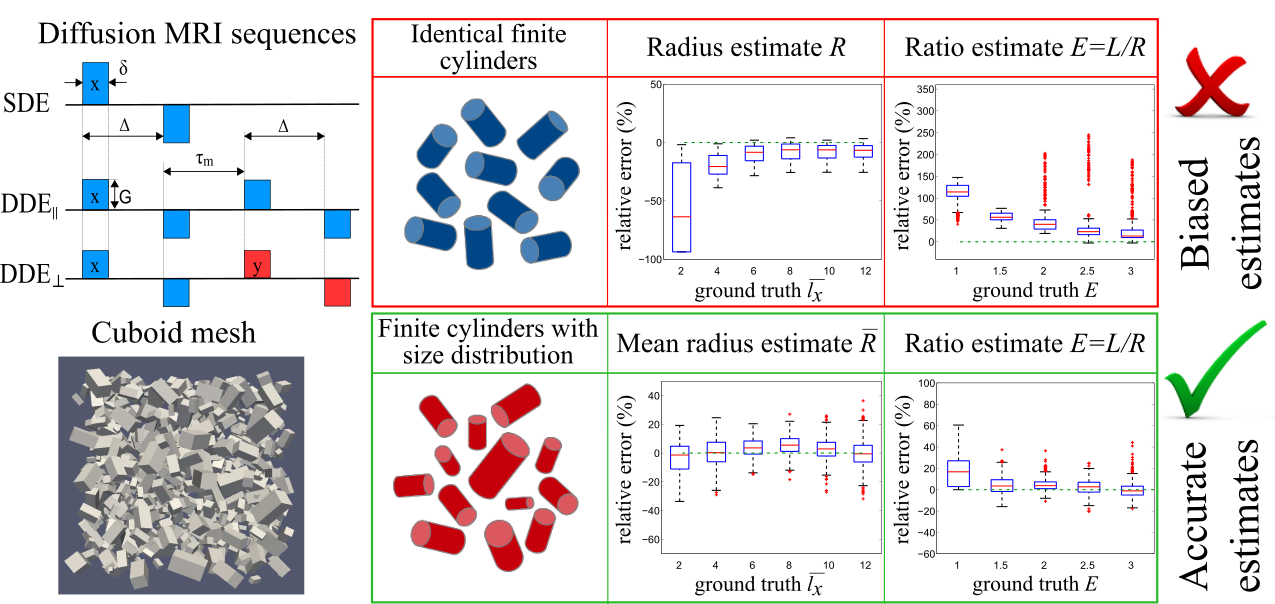
- Personnel: Andrada Ianus.
- Summary: In prostate cancer, the tissue structure changes significantly as the tumour becomes more aggressive. The aim of this project is to develop advanced diffusion MRI techniques, which are sensitive enough to provide a non-invasive classification of tumour grades. The work involves the design of better acquisition sequences, as well as microstructural models for data analysis that capture the changes in cellular architecture.
1.5 Prostate Cancer UK. “CombIning advaNces in imagiNg with biOmarkers for improVed diagnosis of Aggressive prosTate cancEr - INNOVATE”. 1/01/2016 - 31/12/2017.
- Personnel: Hayley Whittaker (PI), Shonit Punwani, Laura Panagiotaki, Elisenda Bonet-Carne, David Atkinson, Edward Johnston, Dave Hawkes, Daniel Alexander et al.
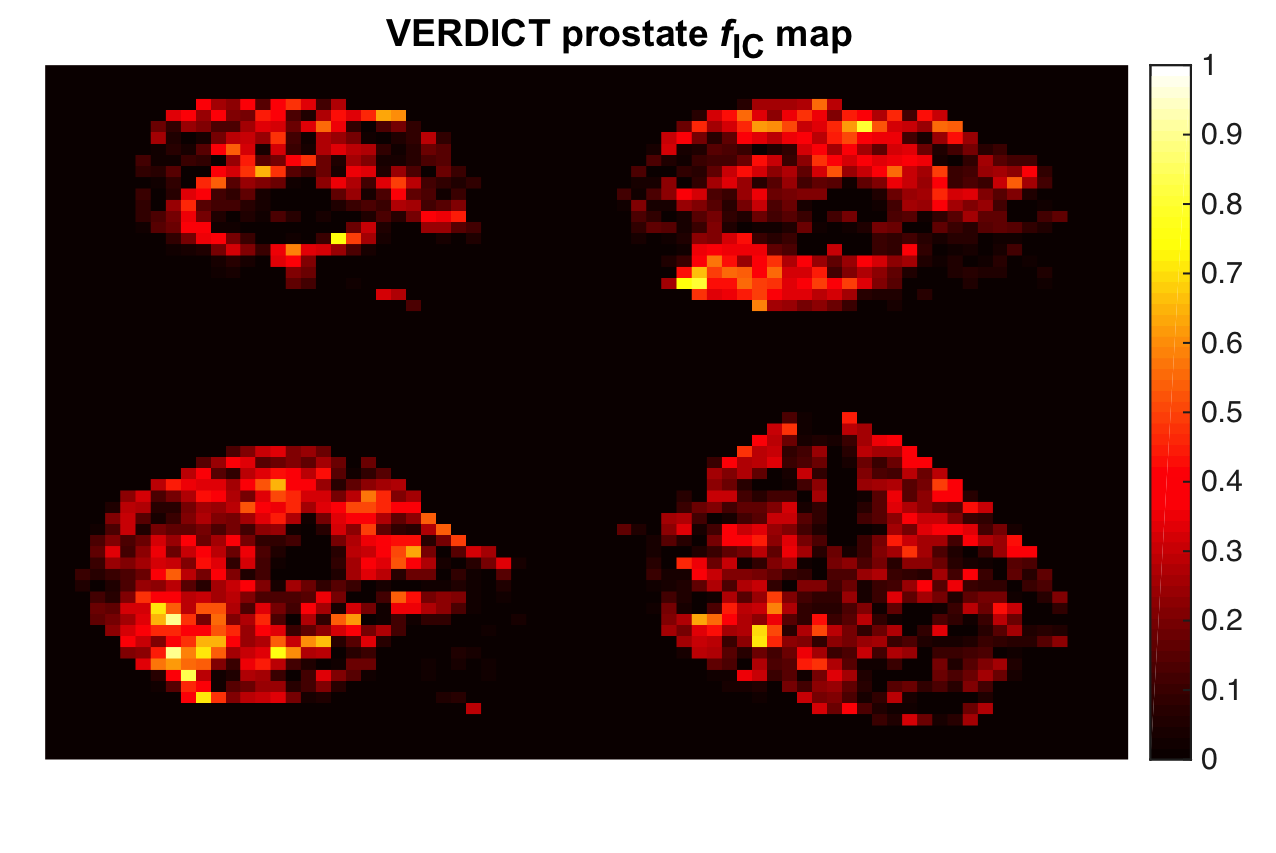
- Summary: INNOVATE is a prospective single centre cohort study in 365 patients which aims to improve the early detection for the aggressive prostate cancer and to increase the MRI diagnostic performance. MIG’s role focuses on testing whether the performance of the multi parametric MRI can be improved by the addition of VERDICT. VERDICT is an advanced diffusion-weighted MRI technique that was designed to capture the main microstructural properties of cancerous tissue. It uses a biophysical model to characterise tissue microstructure. Here is the clinical study record detail website.
- Related Publications:
- Panagiotaki E, Chan RW, Dikaios N, Ahmed HU, Urol F, Callaghan JO, Freeman A, Atkinson D, Punwani S, Hawkes DJ, Alexander DC. Microstructural Characterization of Normal and Malignant Human Prostate Tissue With Vascular , Extracellular , and Restricted Diffusion for Cytometry in Tumours Magnetic Resonance Imaging. Invest Radiol. 2015;50(4):218–27.
- Johnston EW, Pye H, Bonet-Carne E, Panagiotaki E, Patel D, Galazi, Heavey S, Carmona L, Freeman A, Trevisan G, Alexander D.C et al. INNOVATE: A prospective cohort study combining serum and urinary biomarkers with novel diffusion-weighted magnetic resonance imaging for the prediction and characterization of prostate cancer. BMC Cancer 31 Oct 2016 .
- Bonet-Carne E, Daducci A, Panagiotaki E, Johnston E, Stevens N, Atkinson D, Shonit Punwani S, Alexander DC. Non-invasive quantification of prostate cancer using AMICO framework for VERDICT MR. Intl Soc Mag Reson Med 24. Singapore; 2016.
1.6 Microsoft Research PhD Studentship. “Image Quality Transfer”. 1/10/2015 – 28/09/2019.
- Personnel: Ryutaro Tanno (UCL), Aurobrata Ghosh (UCL), Daniel Alexander (UCL), Antonio Criminisi (MSR).
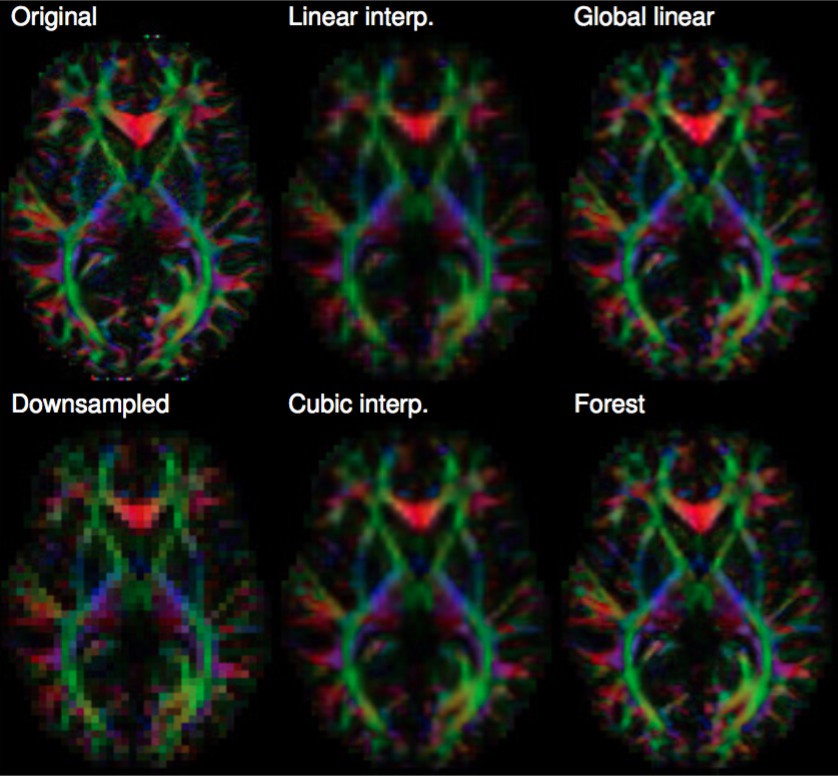
- Summary: This project aims to develop an image reconstruction technology called image quality transfer into a working software tool cutting across several applications. The technique learns a model of the low-level structure of an image ensemble from high quality data sets that are expensive to obtain and uses the model to enhance reconstruction from sparse data that is more practical to acquire. The figure shows a demonstration using MRI data from the human connectome project (HCP) and a simple regression model using random forests to learn a mapping from low-resolution to high-resolution image patches. The idea has important potential in that domain. First, it potentially reveals subtle information hidden in low-resolution images providing more sensitive markers of disease. Second, it potentially supports exploitation of large bodies of historical data from heterogeneous acquisition protocols, which hugely increases statistical power in population or phenotype studies.
- Related Publications:
- Alexander, D. C., Zikic, D., Zhang, J., Zhang, H., & Criminisi, A. "Image quality transfer via random forest regression: applications in diffusion MRI." Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014. Springer International Publishing, 2014. 225-232.
1.7 NIH. “Structure and Function of the Placenta from Implantation to Delivery. A Next Generation MRI Approach”. 17/09/2015 - 31/08/2018.
- Personnel: Paddy Slator, Daniel Alexander, Jana Hutter (KCL), Jo Hajnal (KCL), Mary Rutherford (KCL).
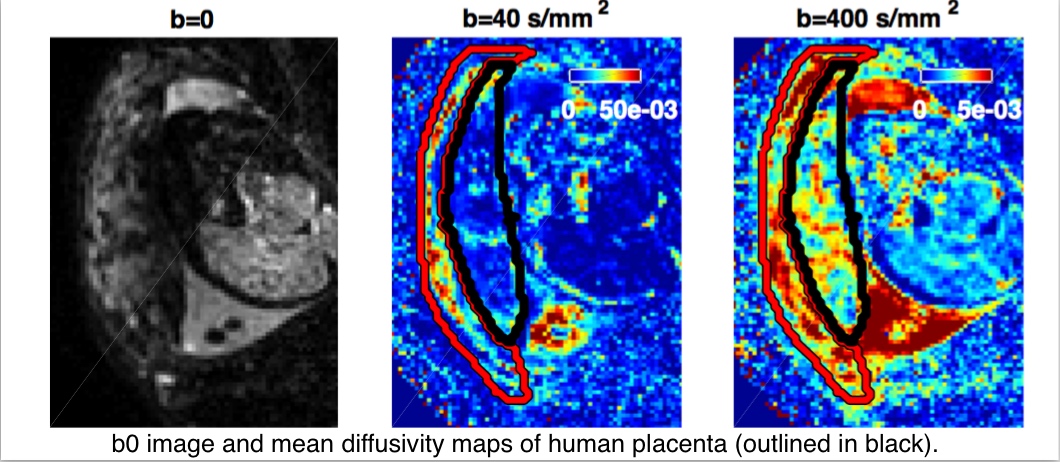
- Summary: Many pregnancy complications stem from placental abnormalities. This project aims to develop tools which assess placental development in the earliest stages of pregnancy. We are developing diffusion-weighted MRI techniques, similar to the NODDI and VERDICT methods, to extract detailed microstructural information from in-vivo human placentae. More details here: http://placentaimagingproject.org/project/.
1.8 EU Horizon 2020. “A Clinical Decision Support system based on Quantitative multimodal brain MRI for personalized treatment in neurological and psychiatric disorders (CDS-QUAMRI)”. 1/09/2015 – 01/09/2020.
- Personnel: Francesco Grussu, Enrico Kaden, Daniel Alexander, Claudia Wheeler-Kingshott
- Summary: The objective of this project is the development of a clinical decision support system for neurological and psychiatric disorders that is based on multimodal quantitative magnetic resonance imaging, advanced feature extraction and multi-parametric classification. To that the quantitative analysis of structural, functional and metabolic MRI data (11 modalities) shall be fully integrated into a single software framework for the first time; support of large data, interoperability and access for non-expert users shall be enabled and a machine learning based classification module shall be developed. The quantification and feature extraction algorithms for metabolic, perfusion, diffusion and functional imaging shall be enhanced to access the full information content of the data independent of vendor specific scan protocols as required for future use in diagnostics, stratification and monitoring of patients.
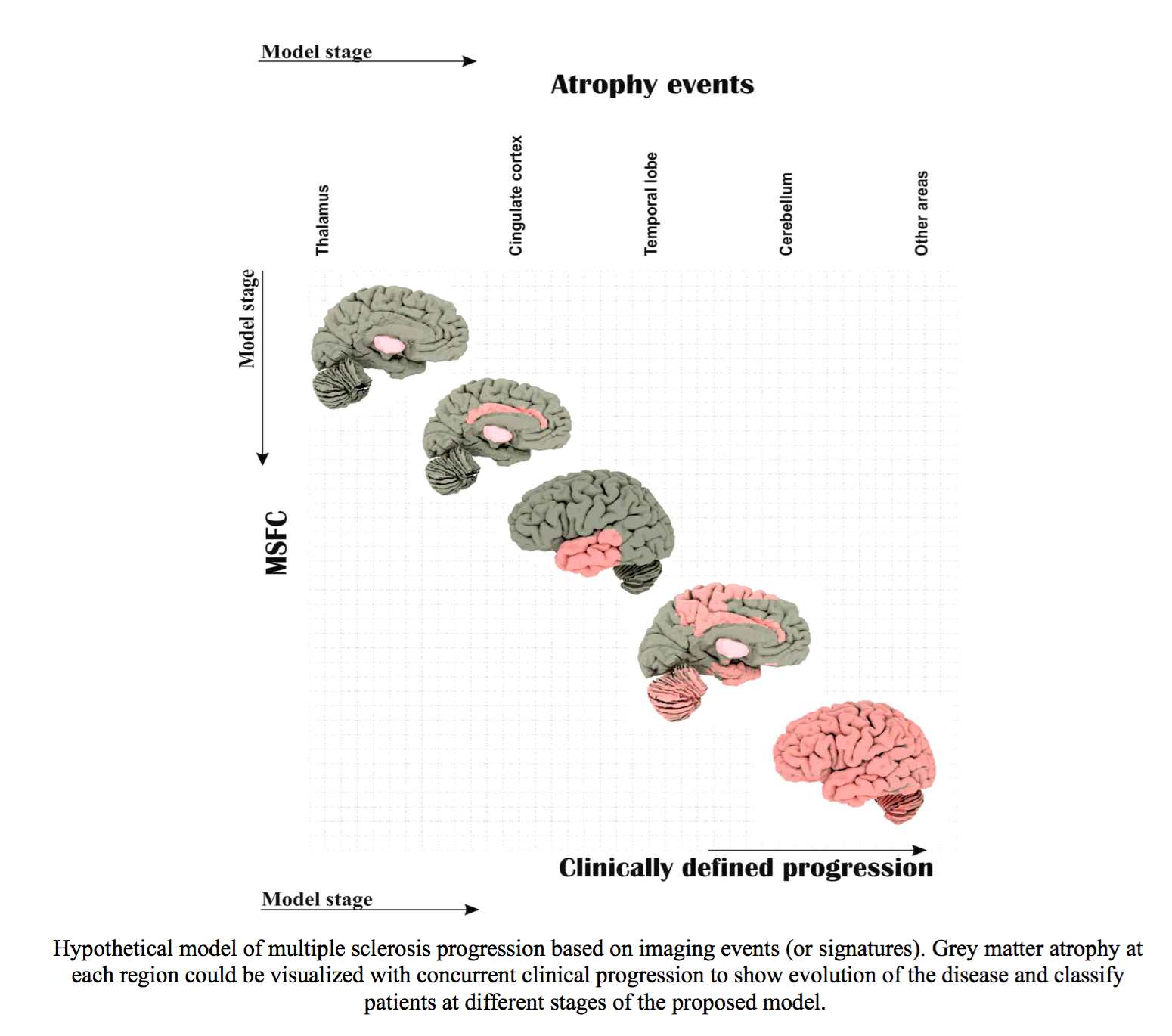
1.9 Magnetic Resonance Imaging in Multiple Sclerosis (MAGNIMS). “To explore imaging signature of multiple sclerosis phenotypes”. 1/07/2015 – 01/07/2016.
- Personnel: Arman Eshaghi, Alan Thompson, Daniel Alexander, Frederik Barkhof, Olga Ciccarelli.
- Summary: Clinicians classify patients with multiple sclerosis (MS) into different phenotypes according to clinical presentation, qualitative MRI and their changes. MS phenotypes are the cornerstone for treatment and rehabilitation strategies, however, clinical classification of patients is crude. We aimed to use a large MRI dataset from different European Multiple Sclerosis Centres (part of MAGNIMS) in London, Rome, Siena, and Amsterdam to explore signatures of different MS phenotypes. Computational modelling of disease progression (such as brain shrinkage) can achieve objective classification of patients. In this project Institute of Neurology works closely with the POND/MIG group.
1.10 EPRSC. “National Facility for In Vivo MR Imaging of Human Tissue Microstructure”. 1/06/2015 – 31/05/2020.
- Personnel: Derek Jones (Cardiff), Richard Bowtell (Nottingham), Geoff Parker (Manchester), Karla Miller (Oxford), Krish Singh (Cardiff), Daniel Alexander (UCL), Hywel Thomas (Cardiff), Flavio Dell'Acqua (KCL), Mara Cercignani (Sussex), Richard Wise (Cardiff).
- Summary: This equipment grant puts in place the National Microstructure Imaging Facility in Cardiff's CUBRIC centre. The device has very high magnetic gradient field strength (up to 300mT/m) enabling exquisite sensitivity to tissue microstructure. It supports the development of future microstructure imaging techniques putting them in place for when this kind of hardware becomes more widespread in the future.
1.11 EPSRC. “Anatomy driven brain connectivity mapping”. 1/06/14 – 31/05/17.
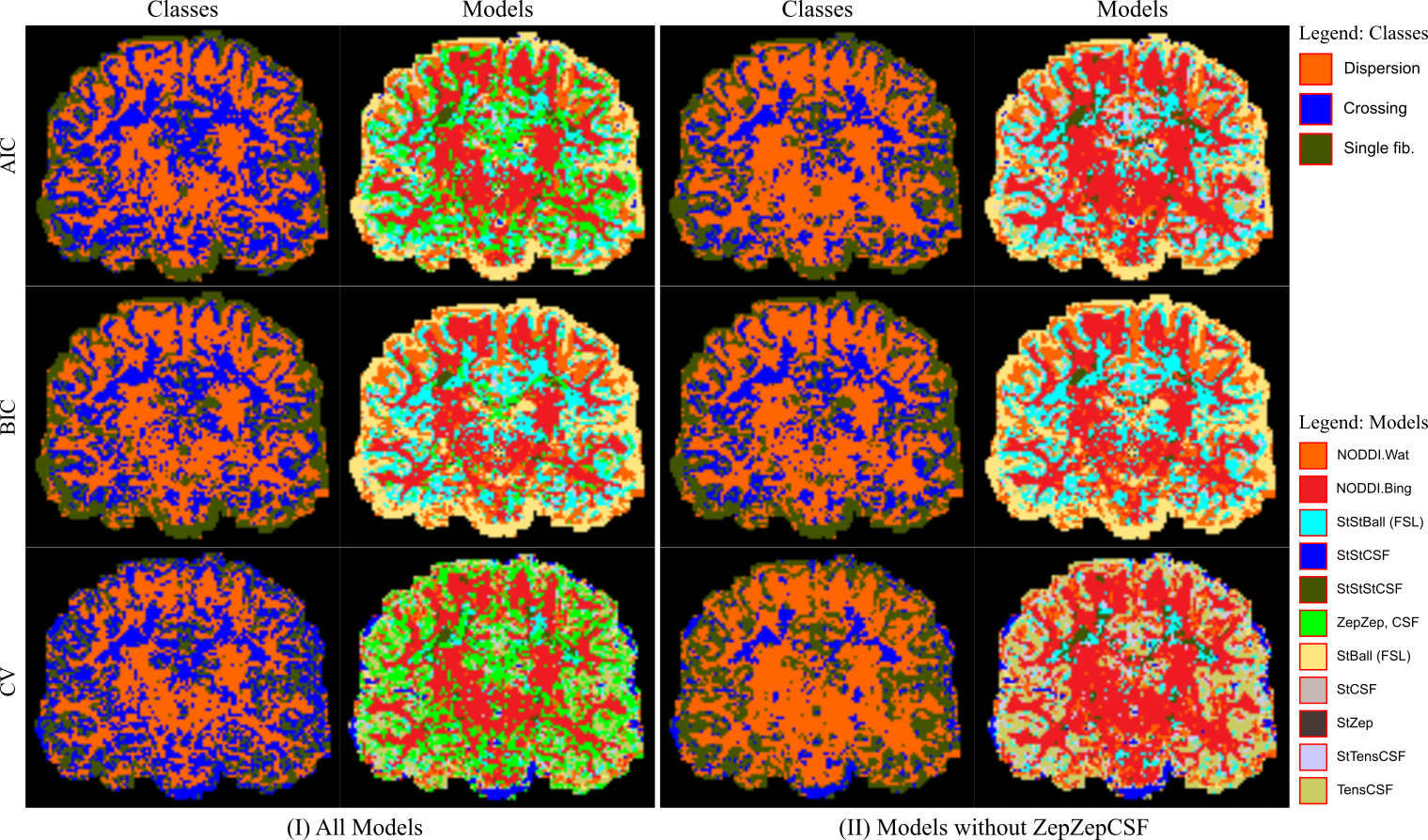
- Personnel: Aurobrata Ghosh, Gary Zhang, Daniel Alexander, Stam Sotiropoulos, Saad Jbabdi, Tim Behrens
- Summary: This project is a collaboration with the FMRIB in Oxford and combines cutting-edge microstructure imaging with prior knowledge learned from histology to bypass inherent limitations in current tractography algorithms. Here is the project outline on the EPSRC website.
- Selected Publications:
- Aurobrata Ghosh, Daniel C. Alexander, and Hui Zhang. Crossing versus Fanning: Model Comparison Using HCP Data. Proceedings of Computational Diffusion MRI MICCAI Workshop, October 2015, Munich, Germany.
1.12 MRC. “In vivo microstructural neuroimaging in infants at risk of developing neurocognitive delay or neurobehavioural disorders”. 1/04/14 – 31/03/19.
- Personnel: Serena Counsell, Jo Hajnal, David Edwards, Paul Aljabar, Gary Zhang, Daniel Alexander.
- Summary: This adapts the latest brain microstructure imaging techniques coming out of MIG for perinatal imaging and exploits the techniques to predict downstream functional deficits.
1.13 UCL CS Research Excellence Studentship. "Automatic analysis of histology with machine learning". 10/03/2014 — 01/09/2017.
- Personnel: Joseph Jacobs, Laura Panagiotaki, Daniel Alexander.
- Summary: This project aims to explore methods for automatic and quantifiable analysis of histological images. Specifically, we intend to develop novel machine learning and pattern recognition for detecting and grading prostate cancer in histology. These methods could then be used to enable computer-aided diagnosis of prostate cancer in histology or for validation of non-invasive imaging modalities.
- Selected Publications:
- Jacobs JG, Panagiotaki E, Alexander DC. Gleason Grading of Prostate Tumours with Max-Margin Conditional Random Fields. MICCAI Workshop on Machine Learning in Medical Imaging 2014.
- Jacobs JG, Johnston E, Freeman A, Patel D, Rodriguez-Justo M, Atkinson D, Punwani S, Brostow G, Alexander DC, Panagiotaki E. Histological validation of VERDICT cellularity map in a prostatectomy case. Intl Soc Mag Reson Med 24. Singapore; 2016.
1.14 UCL SLMS Grand Challenge Studentships project 2013. “Parcellation of the human cortex using quantitative MRI”. 1/10/2013—30/09/2016.

- Personnel: Tara Ganepola, Zoltan Nagy, Daniel C. Alexander and Martin Sereno.
- Summary: The project aims to develop a novel technique for in-vivo cortical parcellation. The approach is based on probing cytoarchitectonic variations in grey matter regions using diffusion MRI and consists of optimising three main aspects of the pipeplie. i) Image acquisition in order to maximise information gain between grey matter regions ii) Methods for describing the underlying tissue microstructure captured in HARDI data and validation of these approaches using ex-vivo data. iii) Determination of appropriate classification methods for this application. The project is a collaboration between the Neuroimaging Group at the Department of Cognitive, Perceptual and Brain Sciences (UCL) and MIG and is supervised by Prof. Martin I. Sereno and Prof. Daniel C. Alexander.
- Selected Publications:
- Zoltan Nagy, Daniel C. Alexander, David L. Thomas, Nikolaus Weiskopf, Martin I. Sereno (2013). Using high angular resolution diffusion imaging data to discriminate cortical regions. PLos One Vol.8 .
- Zoltan Nagy, Tara Ganepola, Martin I. Sereno, Nikolaus Weiskopf, Daniel C. Alexander. Combining HARDI datasets with more than one b-value improves diffusion MRI-based cortical Parcellation. Proceedings of the joint annual meeting ISMRM-ESMRMB 2014, p.0800.
1.15 EPSRC Breast cancer project. “Medical imaging markers of cancer initiation, progression and therapeutic response in the breast based on tissue microstructure”. 1/01/13 – 31/12/15.
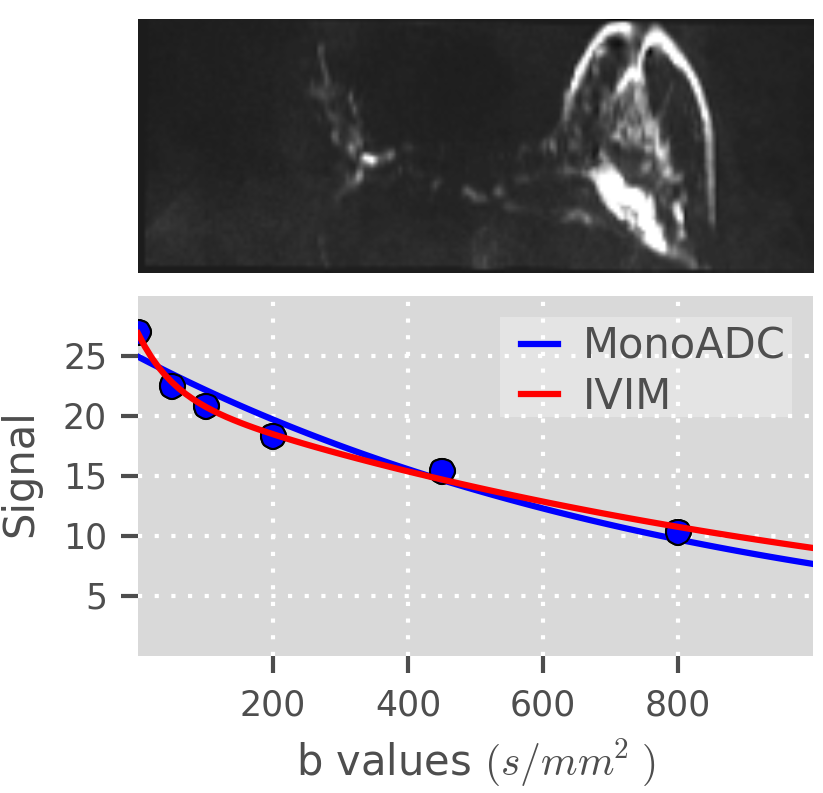
- Personnel: Colleen Bailey, Eleftheria Panagiotaki, Daniel Alexander
- Summary: This project aims to improve breast cancer treatment by searching for imaging features that predict whether patients require therapy and which patients are likely to respond or be resistant. The project is led by Prof Dave Hawkes (CMIC director) and is a collaboration with Ninewells Hospital, Dundee. Data are acquired from many imaging modalities; MIG’s role focuses on developing a model to fit the diffusion MRI signal that reflects the microstructure of the breast tissue. Here is the project outline on the EPSRC website.
- Selected Publications:
- Bailey C, et al. (2014). Modelling vascularity in breast cancer and surrounding stroma using diffusion MRI and intravoxel incoherent motion. LNCS Vol. 8539.
- Bailey C, et al. (2016). Microstructural models for diffusion MRI in breast cancer and surrounding stroma: an ex vivo study. NMR in Biomed.
1.16 UCL SLMS Grand Challenge Studentship project. "Development and validation of machine learning techniques to facilitate diagnosis and predict prognosis in patients with Multiple Sclerosis". 16/11/12-15/11/15
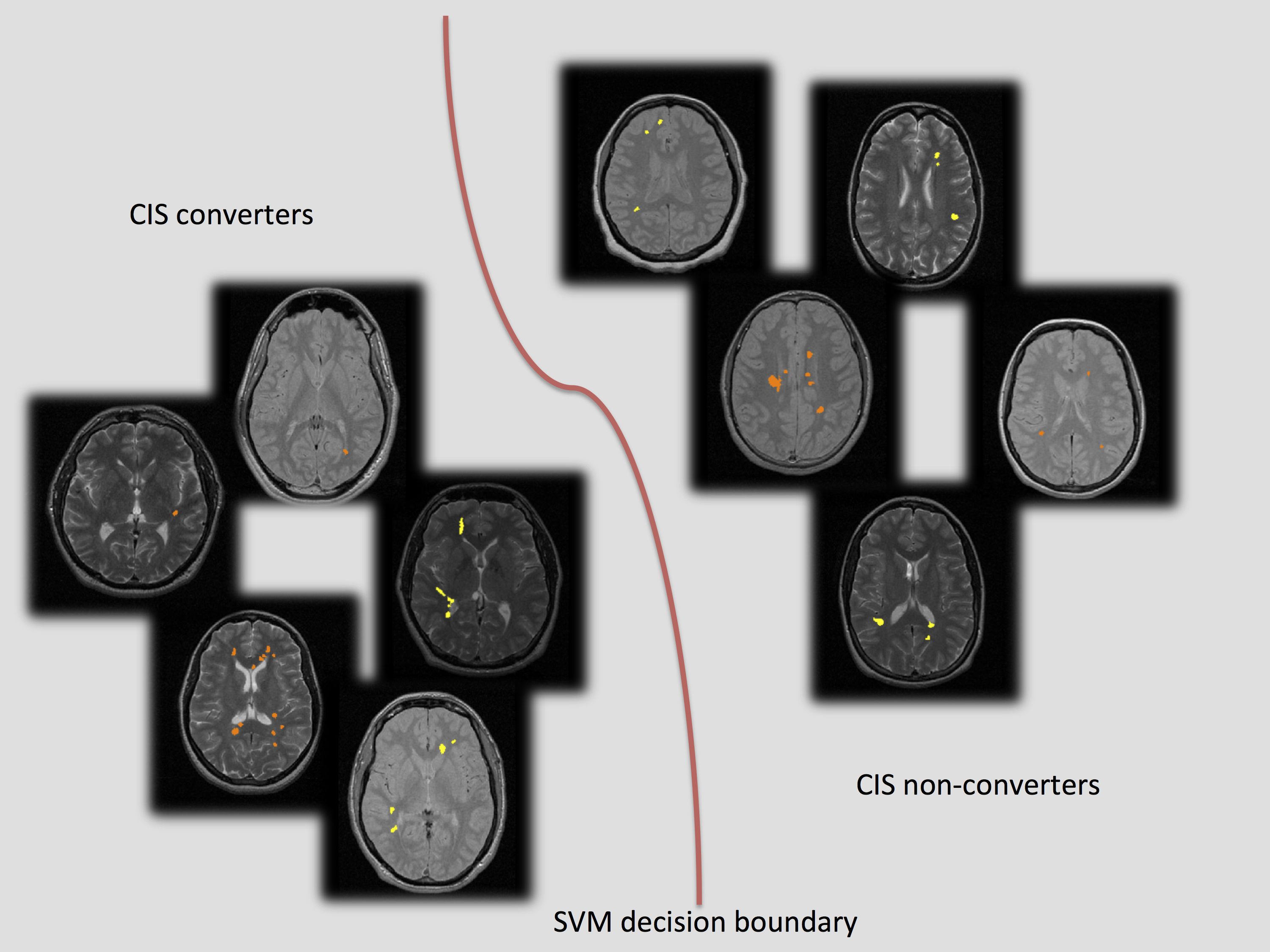
- Personnel: Viktor Wottschel, Daniel Alexander and Olga Ciccarelli.
- Summary: This projects aims to predict diagnosis and prognosis of MS patients from previously aquired MRI data sets. Supervised machine learning classifiers will be explored and optimised to extract information from multi-centre MRI data and create a general model applicable to previously unseen patients. The project is part of a collaboration between the NMR Research Unit at the Insitute of Neurology and MIG and is supervised by Dr Olga Ciccarelli and Prof. Daniel C. Alexander.
- Selected Publications:
- Olga Ciccarelli, et al. (2012). Predicting clinical conversion to multiple sclerosis in patients with clinically isolated syndrome using machine learning techniques, ECTRIMS, No. 113.
- Viktor Wottschel, et al. (2013). Prediction of second neurological attack in patients with clinically isolated syndrome using support vector machines. Proceedings of the international workshop on PRNI 2013.
2. Completed Projects
2.1 CMIC Platform Grant (pump-priming Award). “Enabling multi-site high precision spinal cord MRI”. 01/06/2017 – 01/06/2018.
- Personnel: Francesco Grussu, M. Jorge Cardoso, Claudia A. M. Gandini Wheeler-Kingshott, Daniel C. Alexander.
- Summary: The aim of this project is to adapt recent advances made in the post-processing of diffusion MRI data of the brain to the particular challenges of quantitative MRI (qMRI) of the spinal cord, such as diffusion and myelin imaging. We aim to demonstrate, for the first time, the relevance of combining state-of-the-art noise and Gibbs ringing removal to enhance the reproducibility and the sensitivity to pathology of multi-modal spinal cord qMRI. For this purpose, multi-vendor, multi-modal qMRI data of the spinal cord acquired with cutting edge sequences implemented in clinical scanners will be analysed. The project is a collaboration between MIG, TIG and the UCL Queen Square MS Centre.
2.2 UCL SLMS Grand Challenge Studentships project 2012. “Axonal density as MR imaging biomarker: from bench to bedside”. 1/10/2012—30/09/2015.
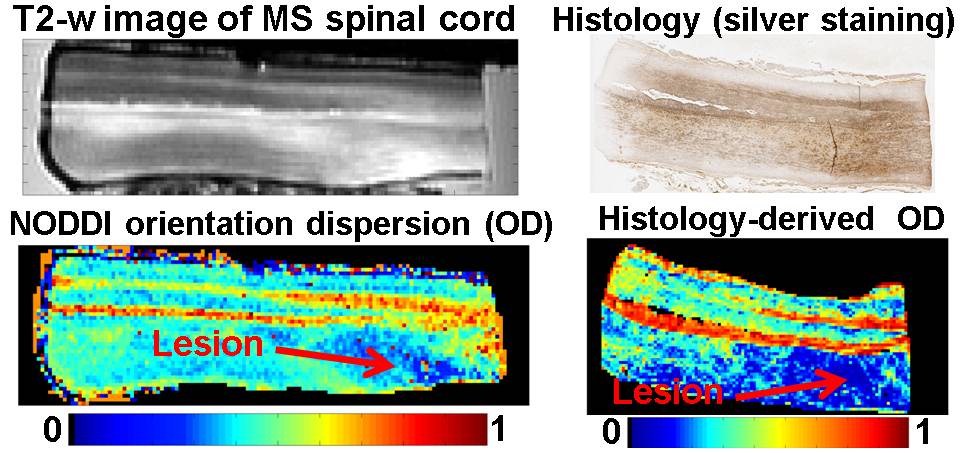
- Personnel: Francesco Grussu, Torben Schneider, Hui Zhang, Daniel C. Alexander and Claudia A. M. Wheeler-Kingshott.
- Summary: the project aims to take advanced diffusion MRI methods for microstructure quantification from the development stage to the clinical application in the spinal cord. The work consists of two main goals: i) appropriate acquisition protocols are to be designed and tested for clinical settings at 3T; ii) the novel diffusion MRI indices are to be compared to histological correlates, in both healthy and multiple sclerosis spinal cord samples, for validation. The project focusses on the optimisation and validation of Neurite Orientation Dispersion and Density Imaging (NODDI) and is part of a collaboration between the NMR Research Unit at the Department of Neuroinflammation (UCL Insitute of Neurology) and MIG. It is supervised by Dr Claudia A. M. Wheeler-Kingshott and Prof. Daniel C. Alexander.
- Selected Publications:
- Grussu F, Schneider T, Yates RL, Zhang H, Gandini Wheeler-Kingshott CAM, DeLuca GC and Alexander DC. A framework for optimal whole-sample histological quantification of neurite orientation dispersion in the human spinal cord. Journal of Neuroscience Methods, Volume 273, 1 November 2016, Pages 20–32.
- Grussu F, Schneider T, Zhang H, Alexander DC, Wheeler-Kingshott CAM. Neurite orientation dispersion and density imaging of the healthy cervical spinal cord in vivo. NeuroImage, Volume 111, 1 May 2015, Pages 590–601.
- Grussu F, Schneider T, Yates RL, Tachrount M, Newcombe J, Zhang H, Alexander DC, DeLuca GC and Wheeler-Kingshott CAM. Histological metrics confirm microstructural characteristics of NODDI indices in multiple sclerosis spinal cord. Proc of ISMRM 2015, p0909, Magna cum Laude award.
2.3 EPSRC Brain connectivity project. “Robust graph analysis of brain connectivity”. 1/06/2012—31/05/2015.
- Personnel: Fani Deligianni, Chris Clark, David Carmichael, Daniel Alexander, Jon Clayden
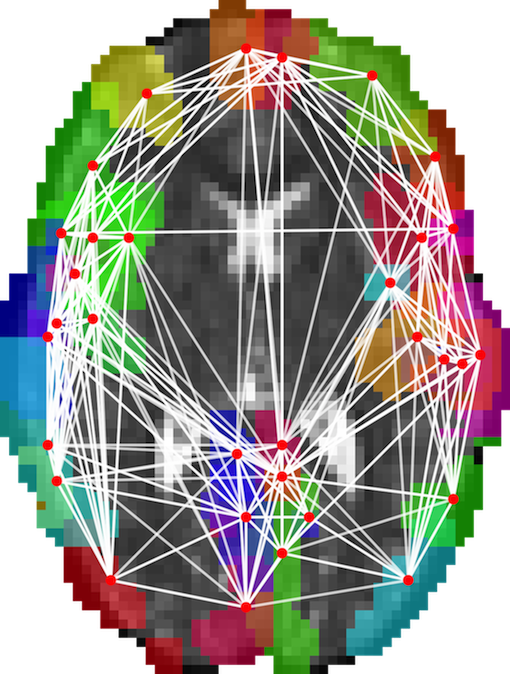
- Summary: This project develops techniques for modelling functional and structural connectivity of the brain using diffusion MRI and EEG. It uses graph analysis to extract salient features and develops statistical techniques for performing group studies of graph-based metrics. Here is the project outline on the EPSRC website.
2.4 Leverhulme Trust Fellowship. "New imaging techniques to reveal the relationship between brain tissue microstructure and brain function". 1/05/2012-31/12/2015.
- Personnel: Ivana Drobnjak
- Summary: This projects aims to develop advanced non-invasive imaging techniques to enable the parallel study of brain microanatomy and function in living humans. The optimisation methods developed in project EP/E007748 produce novel pulse sequences with complex gradient waveforms virtually impossible to devise without the novel algorithmic approach. Phase 1 of this project will implement, refine and validate those sequences on standard clinical MRI systems. Phase 2 will apply the novel sequences to study the microstructure-function relationship in specific neuroscience experiments. The ageing brain will be the principal model for this research, but the new methodology can also be readily applied to study diseases (e.g. Alzheimer’s, Multiple Sclerosis), effects of treatment, or cognitive development.
- Selected Publications:
- Drobnjak I, Zhang H, Ianus A, Kaden E, Alexander D.C. PGSE, OGSE, and sensitivity to axon diameter in diffusion MRI: Insight from a simulation study. Magnetic Resonance in Medicine, Volume 75, February 2016, Pages 688–700.
- Siow B, Drobnjak I, Ianus A, Christie I, Lythgoe M, Alexander D.C. Axon radius estimation with Oscillating Gradient Spin Echo (OGSE) Diffusion MRI. Diffusion Fundamentals, Volume 18(1), 2013, Pages 1-6.
- Drobnjak I, Cruz G, Alexander D.C Optimising oscillating waveform-shape for pore size sensitivity in diffusion-weighted MR. Microporous and Mesoporous Materials, Volume 178, 2013, Pages 11-14.
2.5 EPSRC Healthcare Partnerships Project. "Axon and myelin damage assessed using advanced diffusion imaging: from mathematical models to clinical applications". 1/09/2011-31/08/2014.
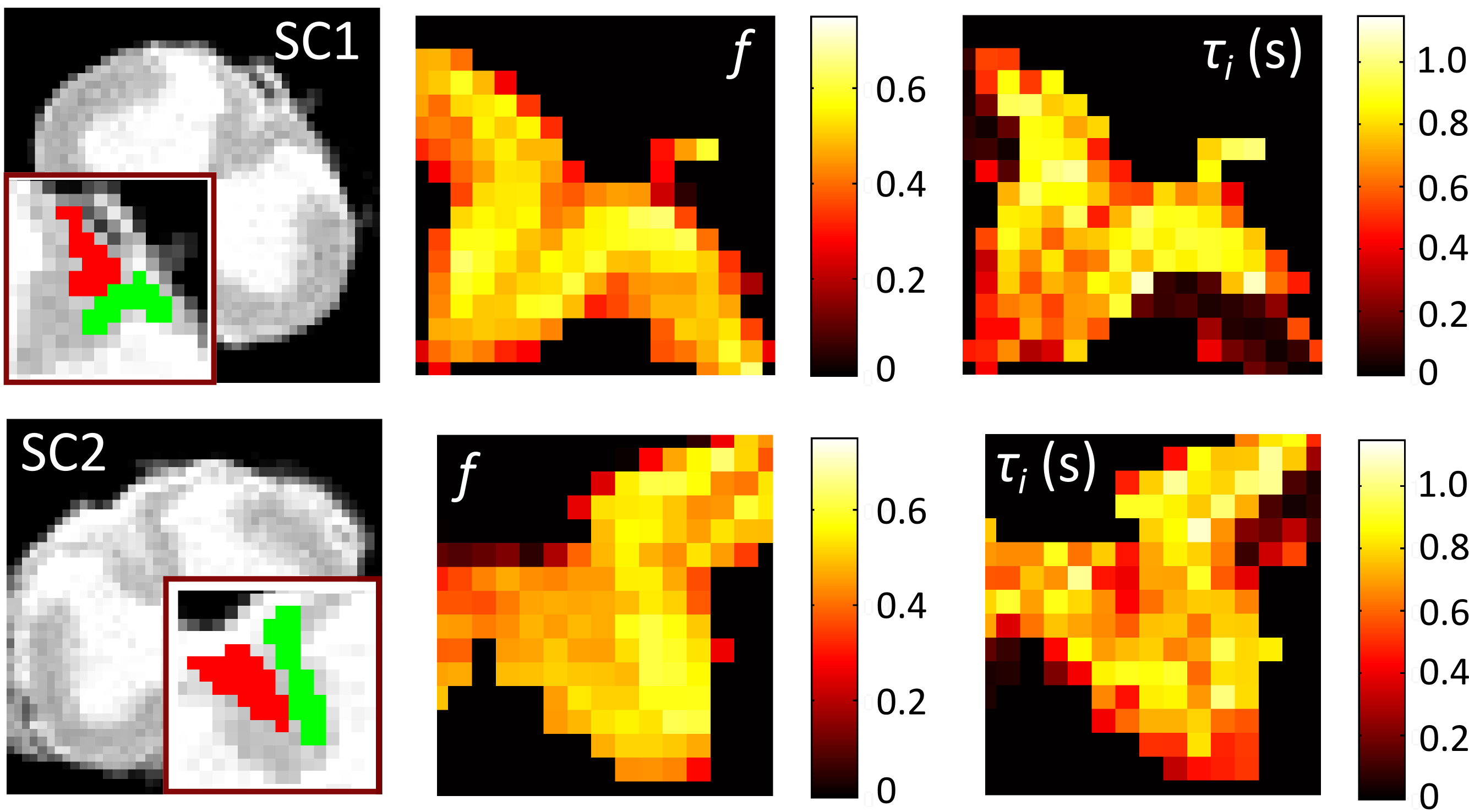
- Personnel: Gemma Nedjati-Gilani, Torben Schneider, Bernard Siow, Uran Ferizi, Francesco Grussu, Daniel Alexander, Claudia Wheeler-Kingshott
- Summary: This project aims to translate emerging ideas in microstructure imaging to the clinic in the specific application of multiple sclerosis diagnosis and imaging. MS has a profound effect on the microstructure of white matter tissue. The NMR unit at UCL's institute of neurology lead this project. Their expertise in clinical MRI combined with MIG's mathematical and computational modelling techniques enable development of new MRI modalities tuned for specific sensitivity to MS pathology. See the project website for more details.
- Selected Publications:
- U. Ferizi, T. Schneider, T. Witzel, L.L Wald, H. Zhang, C. A. M. Wheeler-Kingshott and D.C. Alexander (2015). White matter compartment models for in vivo diffusion MRI at 300mT/m. data. Neuroimage, Vol 118 pp. 468-483, 2015.
- Nedjati-Gilani, G.L. et al (2014). Machine learning based compartment models with permeability for white matter microstructure imaging, MICCAI, pp 257-264.
- Ferizi, U. et al (2014). A ranking of diffusion MRI compartment models with in vivo human brain data, Magnetic Resonance in Medicine, Vol 72, pp 1785-92.
- Ferizi, U. et al (2013). The importance of being dispersed: A ranking of diffusion MRI models for fibre dispersion using in vivo human brain data, MICCAI, pp 74-81.
2.6 EPSRC Program grant. “Intelligent Imaging: Motion, Form and Function Across Scale”. 1/06/2010—31/05/2015.
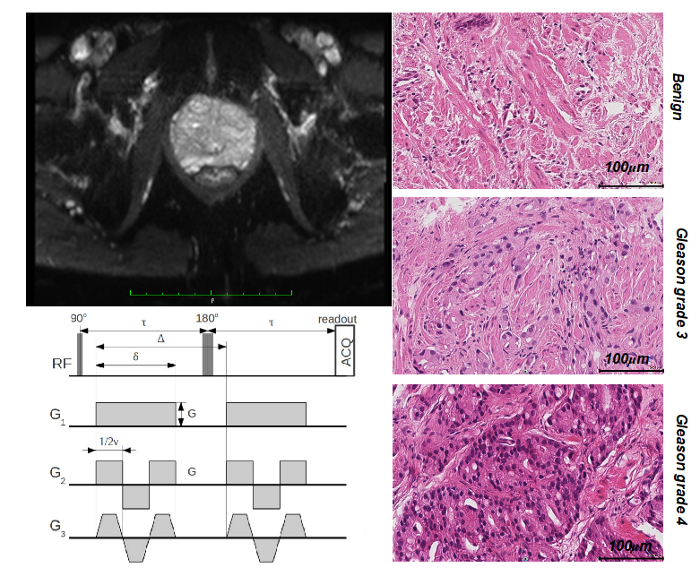
- Personnel: Andrada Ianus, Eleftheria Panagiotaki, Daniel Alexander
- Summary: This Program Grant is a collaboration between UCL-CMIC, King's College London and Imperial to target clinical information obtainable from imaging devices directly without necessarily reconstructing images as an intermediate step. Prof Dave Hawkes (CMIC director) leads the project. MIG's role is modelling normal and pathological tissue structure in the prostate and developing imaging techniques to probe them non-invasively.
- Selected Publications:
- E. Panagiotaki, R.W. Chan, N. Dikaios, J. O'Callaghan, A. Freeman, D. Atkinson, S. Punwani, D.J. Hawkes, and D.C Alexander Microstructural characterisation of normal and malignant human prostate tissue with VERDICT MRI. Investigative Radiology, 50(4), 218–227, 2015.
- Panagiotaki, E., Walker-Samuel, S., Siow, B., Johnson, P., Rajkumarc, V., Pedley, R., Lythgoe,, Alexander, D. C. (2014). Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Research 2014;74 (7) 1902-1912 doi:10.1158/0008-5472.CAN-13-2511.
- Ianus, A., Siow, B., Drobnjak, I., Zhang, H. and Alexander, D. C. (2013). Gaussian phase distribution approximations for oscillating gradient spin echo diffusion MRI. Journal of Magnetic Resonance 227, pp.25-34.
- Ianus, A., Drobnjak, I., and Alexander, D. C. (2016). Model-based estimation of microscopic anisotropy using diffusion MRI: a simulation study. NMR in Biomedicine, accepted.
2.7 EPSRC Leadership Fellowship. "Direct measurements of microstructure from MRI." EP/E007748. 1/10/2008—30/09/2014.
- Personnel: Bernard Siow, Ivana Drobnjak, Matt Hall, Enrico Kaden, Tingting Wang, Simon Richardson, Daniel Alexander
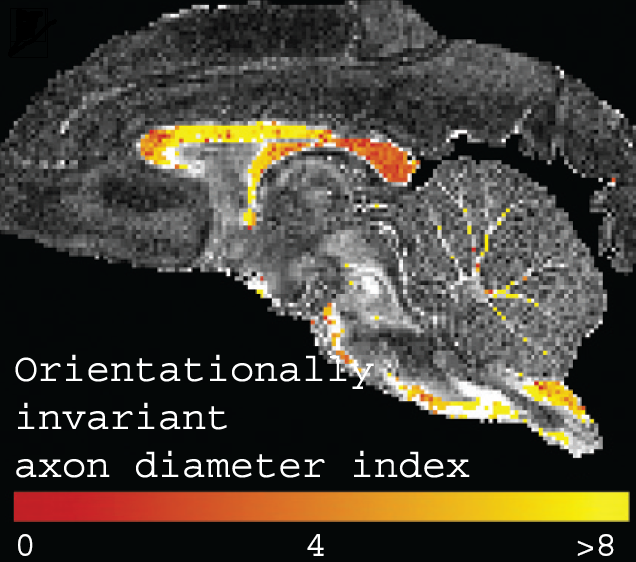
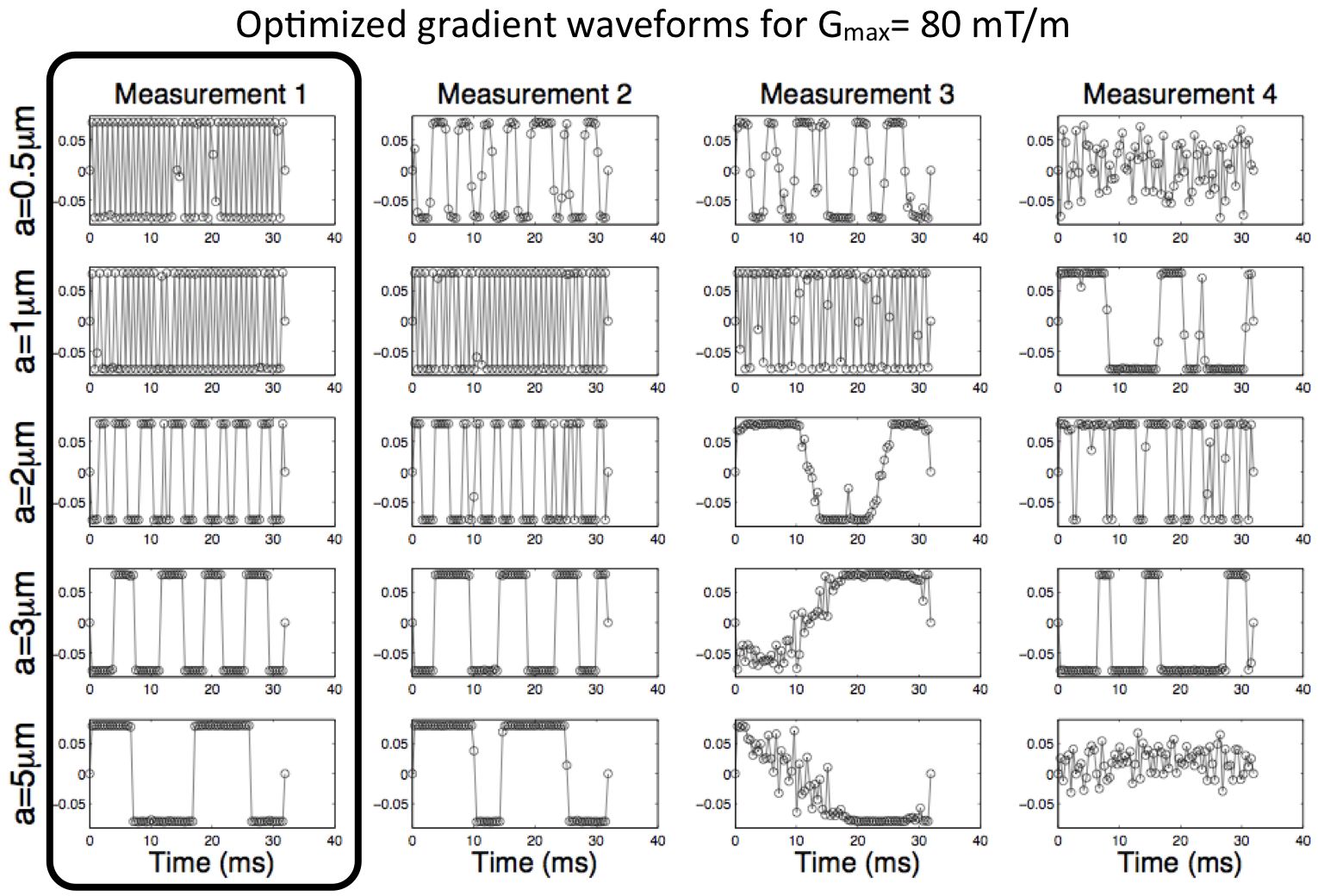
- Summary: This project develops and implements a variety of tissue models for microstructure imaging in the brain. It also develops experiment design optimization techniques that use those models to identify economical imaging protocols that are achievable on live human subjects and patients. A starting point is measuring the diameter of axons in white matter of live human subjects, which other groups have demonstrated in vitro. Later stages will use similar approaches to target specific cellular changes known to occur in particular diseases, such as Alzheimer's disease, epilepsy and multiple sclerosis. The intended output is MRI techniques specifically tuned for the pathologies of those diseases, which should improve diagnosis, prognosis and treatment planning and be valuable for drug trials.
- Selected Publications:
- E. Kaden, F. Kruggel, and D.C. Alexander. Quantitative mapping of the per-axon diffusion coefficients in brain white matter. Magnetic Resonance in Medicine (In Press).
- Richardson, S., Siow, B., Panagiotaki, E., Schneider, T., Lythgoe, M. F., Alexander, D. C.Viable and fixed white matter: diffusion magnetic resonance comparisons and contrasts at physiological temperature Magnetic Resonance in Medicine 72, 1151-61, 2014.
- Richardson, S., Siow, B., Batchelor, A. M., Lythgoe, M. F., Alexander, D. C.A viable isolated tissue system: a tool for detailed MR measurements and controlled perturbation in physiologically stable tissue Magnetic Resonance in Medicine 69, 1603-10, 2013.
- Siow, B., Drobnjak, I., Chatterjee, A., Lythgoe, M. F., Alexander, D. C.Estimation of pore size in a microstructure phantom using the optimised gradient waveform diffusion weighted NMR sequence Journal of Magnetic Resonance 214(1), 51-60, 2012.
- I. Drobnjak, D.C. Alexander Optimising time-varying gradient orientation for microstructure sensitivity in diffusion-weighted MR Journal of Magnetic Resonance 212(2), 344-354, 2011.
- I. Drobnjak, H. Zhang, M.G. Hall, D.C. Alexander The matrix formalism for generalised gradients with time-varying orientation in diffusion NMR Journal of Magnetic Resonance 210, 151-157, 2011.
- I. Drobnjak, B. Siow, D.C. Alexander Optimizing gradient waveforms for microstructure sensitivity in diffusion-weighted MR Journal of Magnetic Resonance 206, 41-51, 2010.
- D.C. Alexander, P.L. Hubbard, M.G. Hall, L.A. Moore, M. Ptito, G.J.M. Parker and T.D. Dyrby Orientationally invariant indices of axon diameter and density from diffusion MRI NeuroImage 54, 1374-1389, 2010.
- Alexander,D.C. (2008). A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magnetic Resonance in Medicine 60, 439-448. ISSN: 0740-3194
2.8 European Commission (Future and Emerging Technologies program). “CONNECT: Consortium of NeuroImagers for the Non-invasive Exploration of Brain Connectivity and Tractography”. 1/01/2010—31/10/2012.
- Personnel: Gary Zhang, Daniel Alexander

- Summary: The CONNECT consortium brings together the very best among the European Union's world-leading diffusion MRI community with the aim to create definitive atlases of the brain connectivity for the healthy developing and adult brain that will serve as a long-lasting reference for the neuroscience and medical community. The completion of this project will fill a critical need for the comprehensive mapping of the connections and tissue microstructure of the human brain. The project is coordinated by Dr. Yaniv Assaf (Tel Aviv University), Prof. Daniel Alexander (UCL), and Prof. Derek K Jones (Cardiff University), and involves 12 research teams in 10 institutes across 7 countries. The sub-aim of the UCL team is in the design and implementation of microstructure imaging and analysis tools and their application to the direct mapping of tissue microstructure of the human brain.
- Selected Publications:
- Lundell H, Alexander DC, Dyrby TB (2014). High angular resolution diffusion imaging with stimulated echoes: compensation and correction in experiment design and analysis. NMR in Biomedicine Vol. 27, pp. 918-25.
- Assaf Y, Alexander DC, Jones DK, Bizzi A, Behrens TE, Clark CA, Cohen Y, Dyrby TB, Huppi PS, Knoesche TR, Lebihan D, Parker GJ, Poupon C; CONNECT consortium, Anaby D, Anwander A, Bar L, Barazany D, Blumenfeld-Katzir T, De-Santis S, Duclap D, Figini M, Fischi E, Guevara P, Hubbard P, Hofstetter S, Jbabdi S, Kunz N, Lazeyras F, Lebois A, Liptrot MG, Lundell H, Mangin JF, Dominguez DM, Morozov D, Schreiber J, Seunarine K, Nava S, Poupon C, Riffert T, Sasson E, Schmitt B, Shemesh N, Sotiropoulos SN, Tavor I, Zhang HG, Zhou FL (2013). The CONNECT project: Combining macro- and micro-structure. Neuroimage Vol. 80, pp. 273-282.
- Alexander DC, Dyrby TB (2013). Diffusion imaging with stimulated echoes: signal models and experiment design. arXiv:1305.7367 [physics.med-ph].
- Dyrby TB, Søgaard LV, Hall MG, Ptito M, Alexander DC (2013). Contrast and stability of the axon diameter index from microstructure imaging with diffusion MRI. Magnetic Resonance in Medicine, Vol. 70, pp. 711-21.
- Zhang H, Schneider, T, Wheeler-Kingshott, CAM, Alexander DC (2012). NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage, Vol. 61(5), pp. 1000-1016 .
- Zhang H, Dyrby T, Alexander DC (2011). Axon diameter mapping in crossing fibers with diffusion MRI. Medical Image Computing and Computer Assisted Intervention (MICCAI), Part II, LNCS 6892, pp 82-89.
- Zhang H, Hubbard P, Parker GJ, Alexander DC (2011). Axon diameter mapping in the presence of orientation dispersion with diffusion MRI. NeuroImage, Vol. 56, pp. 1301-1315.
- Zhang H, Barazany D, Assaf Y, Lundell H M, Alexander D C, and Dyrby T B (2011). A comparative study of axon diameter imaging techniques using diffusion MRI. Proceedings of the International Society of Magnetic Resonance in Medicine (ISMRM) 2011, pp. 83-83.
- Zhang H, Alexander DC (2010). Axon diameter mapping in the presence of orientation dispersion with diffusion MRI. Medical Image Computing and Computer Assisted Intervention (MICCAI), Part I, LNCS 6361, pp 640-647.
- Sherbondy, AJ, Rowe, MC, Alexander DC (2010). MicroTrack: an algorithm for concurrent projectome and microstructure estimation.. Medical Image Computing and Computer Assisted Intervention (MICCAI), Part I, LNCS 6361, pp 183-190
2.9 International Spinal Research Trust (ISRT). "Spinal Cord Diffusion Imaging: Challenging Characterization and Prognostic Value'' Natalie Rose-Barr Studentship. 1/10/2008--30/9/2011.
- Personnel: Torben Schneider, Daniel Alexander, Claudia Wheeler-Kingshott.
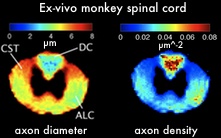
- Summary: Magnetic Resonance Imaging has shown to be very useful in evaluating several aspects of spinal cord injury (SCI) but traditional MRI has limited predictive value because it cannot readily distinguish important fine structural detail within the cord. Diffusion MRI is a very promising technique that is sensitive to microstructural tissue characteristics but it has yet to be applied to the spinal cord. In this project we investigate the potential of existing methods like diffusion tensor imaging and develop new techniques that are more sensitive to specific tissue characteristics like axon diameter and density to help understanding the underlying structural changes in SCI. With new SCI treatment strategies on the horizon, this technique will provide novel biomarkers for measuring therapy outcome and will be of great significance in monitoring the success of future clinical trials.
- Selected Publications:
- Schneider T, Wheeler-Kingshott CAM, Alexander DC. (2010). In-vivo estimates of axonal characteristics using optimized diffusion MRI protocols for single fibre orientation. 13th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI2010).
- Schneider T, Alexander DC, Wheeler-Kingshott CAM. (2010). Optimized diffusion MRI protocols for estimating axon diameter with known fibre orientation. International society for magnetic resonance in medicine (ISMRM). Stockholm. p 1561.
2.10 EPSRC. "Monte Carlo random effects modelling in diffusion MRI: a new window on microstructure and white matter architecture.'' EP/G025452/01. 1/10/2008--30/9/2011.
- Personnel: Martin King, Daniel Alexander, David Gadian, Chris Clark
- Summary: Microstructure imaging is hard, because sensitivity of MRI measurements to interesting features of tissue microstructure, such as axon diameter or the size of cancer cells, is low, particularly in measurements from clinical MRI scanners. This project uses advanced statistical techniques to pool information from similar image voxels and thus increase sensitivity to the subtle changes we look for in microstructure imaging. Early stages of the project use random effects modelling for increasing the accuracy of estimates of crossing fibre estimates. Later stages will use the same approach for mapping axon diameter and density in live human subjects.
- Selected Publications:
- King MD, Gadian DG, Clark CA (2009). A random effects modelling approach to the crossing-fibre problem in tractography. Neuroimage 44(3):753-68.
2.11 EPSRC CASE Studentship with GSK. 28/9/2008--27/9/2011.
- Personnel: Gemma Morgan, Daniel Alexander
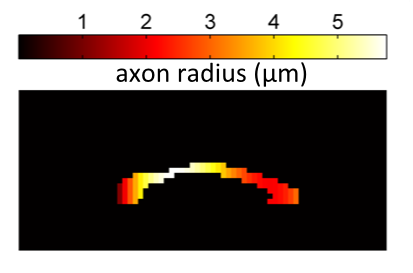
- Summary: Schizophrenia is a severe psychiatric disease which affects approximately 1% of the population. Both structural and functional changes to the brain have been implicated in the disease, although we still have a very limited understanding of what these changes are. Previous structural studies have used DTI to measure changes (often conflicting!) in FA and MD, but these indices are vague and non-specific. In this project, we aim to identify new imaging biomarkers which directly relate to the underlying microstructure and develop techniques for mapping the microstructural changes in schizophrenia in a clinical setting.
- Selected publications:
- Morgan GL, Zhang H, Whitcher B and Alexander DC. (2010). A spatial variation model of white matter microstructure. Workshop on Computational Diffusion MRI, Medical Image Computing and Computer-assisted Intervention (MICCAI)
- Morgan GL, Zhang H, Whitcher B and Alexander DC. (2011). A Bayesian framework for modelling the regional variation of white matter microstructure. Medical Image Understanding and Analysis
2.12 EPSRC Doctoral Prize (PhD+). “Differentiating grades of brain tumour with DW-MRI”. 1/01/2011-1/01/2012.
- Personnel: Eleftheria Panagiotaki, Daniel Alexander
- Summary: The EPSRC Doctoral prize supports the award of fellowships of up to one year's duration. The scheme allows students to advance their PhD research as fellows. The motivation for this project has been the replacement of biopsy with non-invasive imaging to determine brain tumour grade. An accurate classification of the brain tumour grade is imperative as it determines the treatment plan for the patient. However, the standard classification comes from histopathology after the traumatic and uncomfortable biopsy which carries its own risks of mortality. The aim of this research has been to develop models of the diffusion-weighted (DW-MRI) signal in brain tumours ultimately to enable non-invasive differentiation between low and high grades. The plan for this one year was to identify a parsimonious parametric model relating all important microstructure indices, such as cellularity and mitotic index, to the signal. As proof of concept, the project investigated animal models of cancer and brain tumour biopsy samples which reflect the clinical condition.
- Selected Publications:
- Panagiotaki, E., Walker-Samuel, S., Siow, B., Johnson, P., Rajkumarc, V., Pedley, R., Lythgoe,, Alexander, D. C. (2014). Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Research 2014;74 (7) 1902-1912 doi:10.1158/0008-5472.CAN-13-2511.
2.13 EPSRC. "Monte-Carlo simulation framework for diffusion MRI.'' EP/E056938/1. 1/9/07--31/8/10.
- Personnel: Eleftheria Panagiotaki, Matt Hall, Daniel Alexander
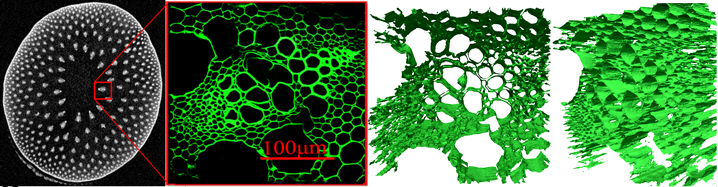
- Summary: A frequent difficulty with new imaging methodologies, particularly imaging in highly complicated structures, is that the lack of a ground truth with which to validate results. This project involves constructing a Monte-Carlo simulation framework for diffusion MRI which is flexible and capable of synthesising data from highly detailed environments. The simulation generates trajectories for a population of diffusing spins in a known environment that restricts their motion as well as simulating the action of an appropriate MR pulse sequence in order to directly generate diffusion attenuated signals.
- A significant part of this project has involved constructing highly detailed triangle-mesh substrates from Confocal Laser Microscopy images of tissue from biological samples. We acquire a stack of images which are then processed into a mesh of a few hundred-thousand triangles which are then imported into the simulation and used to generate signals. We can then compare the simulated signals the results of putting the same sample the MRI scanner. This gives us confidence that the simulation is a capable of closely reproducing data from a complex sample.
- The simulation has been used in many studies of new analysis methods, including several of the other projects on this page. In addition to triangle meshes, it can use cylinders of different radii and packings (ordered and disordered) and is capable of simulating an almost unlimited set of pulse-sequence waveforms.
- Selected Publications:
- Panagiotaki, E., Schneider, T., Siow, B., Hall, M. G., Lythgoe, M. F., Alexander, D. C. (2012). Compartment models of the diffusion MR signal in brain white matter: A taxonomy and comparison. Neuroimage 59(3), 2241-2254 doi:10.1016/j.neuroimage.2011.09.081
- Panagiotaki E, Hall MG, Zhang H, Siow B, Lythgoe MF, Alexander DC. (2010). High-fidelity meshes from tissue samples for diffusion MRI simulations. Medical Image Computing and Computer Assisted Intervention (MICCAI).
- Panagiotaki,E., Fonteijn,H., Siow,B., Hall,M.G., Price,A., Lythgoe,M.F., Alexander,D.C. (2009). Two-compartment models of the diusion MR signal in brain white matter. Medical Image Computing and Computer Assisted Intervention (MICCAI) London:Springer, 1, 329-336
- Hall,M.G., Alexander,D.C. (2009). Convergence and Parameter Choice for Monte-CarloSimulations of Diffusion MRI. IEEE Transactions on Medical Imaging 28(9), 1354-1364. ISSN: 0278-0062
2.14 NHS, Frenchay Hospital. "The optimisation of diffusion imaging to quantify fluid flow within the central nervous system'' 1/1/2008--31/12/2009.
- Personnel: Hubert Fonteijn, Daniel Alexander
- Summary: The delivery of drugs to the brain via normal ways (i.e. intraveneously) is complicated because of the Blood-Brain Barrier. An alternative to intraveneous delivery is Convection Enhanced Delivery (CED). In CED a catheter is placed directly into the brain from which drugs are infused. Drugs either fill up the volume locally, or follow white matter paths to reach more distant brain regions. This might for instance be one or several tumour sites. CED can only be used when we know beforehand where the drugs will end up. The goal of this project is to develop accurate simulations of CED. We use DWI-derived parameters, such as fibre direction, as an input for these simulations and assess their accuracy. We also work on infusion planning algorithms, which determine what the optimal infusion site is given a Region of Interest.
- Selected Publications
- Fonteijn, H.M., Woodhouse, M., White, E., Gill, S.S., Alexander, D.C. Determining infusion sites for Convection Enhanced Delivery using probabilistic tractography. Presented at Computational Diffusion MRI workshop, Medical Image Computing and Computer Assisted Intervention (MICCAI), Beijing, 2010
2.15 EPSRC CASE Studentship. 1/10/2005--31/9/2008.
- Personnel: Tony Shepherd, Daniel Alexander

- Summary: This project developed an interactive segmentation system for outlining regions of interest in images. The application of interest was lesion contouring in medical imaging, but the system has broader potential application. The key technological advance was to devise statistical shape models that do not require shapes with distinctive and consistent features. Experiments demonstrate that modelling the shapes of lesions in this way reduces the interaction time required by users outlining regions of interest.
- Selected Publications:
- Shepherd, T, Prince, SJD, Alexander, DC (2012) Interactive Lesion Segmentation with Shape Priors from Off-line and On-line Learning. IEEE Trans. Medical Imaging, Vol. 31(9), pp. 1698-1712.
- Shepherd,T., Alexander,D.C. (2008). Supervised Methods for Perfect Segmentation in Medical Images. IEEE International Conference on Image Processing, 1460-1463
2.16 EPSRC. "Multiple-fibre reconstruction in diffusion MRI''. GR/T22858/01. 1/1/2005--31/12/2007.
- Personnel: Kiran Seunarine, Matt Hall, Daniel Alexander

- Summary: At the start of this project, multi-fibre reconstruction algorithms (often called HARDI algorithms, although HARDI really refers to the imaging protocol) were just beginning to emerge. The aim was to develop and validate the algorithms to the point where they could be used routinely in tractography or biomedical studies of tissue microstructure. The project implemented a variety of techniques within the Camino toolkit, as well as algorithms for exploiting them in tractography. It produced several review papers on the topic.
- Selected Publications:
- Seunarine,K.K.,Alexander,D.C. (2009) Multiple fibres: beyond the diffusion tensor. Chapter in Diffusion MRI: From quantitative measurement to in-vivo neuroanatomy edited by Johansen-Berg,H. and Behrens,T.E.B., Elsevier.
- Seunarine,K.K., Cook,P.A., Hall,M.G., Embleton K, Parker,G.J., Alexander,D.C. (2007). Exploiting Peak anisotropy for tracking through complex structures. ICCV workshop on Mathematical methods in biomedical image analysis (MMBIA)
- Alexander,D.C. (2005). Maximum entropy spherical deconvolution for diffusion MRI. Information Processing in Medical Imaging, Christensen,G.E., Sonka,M. (ed.) Lecture notes in computer science series. New York:Springer, 3565, 76-87
- Alexander,D.C. (2005). Multiple fibre reconstruction algorithms for diffusion MRI. Annals of the New York Academy of Sciences 1046, 113-133. ISSN: 0077-8923
- Alexander,D.C., Barker,G.J. (2005). Optimal imaging parameters for fibre-orientation estimation in diffusion MRI. NeuroImage 27, 357-367. ISSN: 1053-8119
- Parker,G.J.M., Alexander,D.C. (2005). Probabilistic anatomical connectivity derived from the microscopic persistent angular structure of cerebral tissue. Philosophical Transactions of the Royal Society: B 360(1457), 893-902. ISSN: 0264-3839
- Jansons,K.M., Alexander,D.C. (2003). Persistent Angular Structure: New Insights from Diffusion Magnetic Resonance Imaging Data. Inverse Problems 19, 1031-1046. ISSN: 0266-5611
- Alexander,D.C., Barker,G.J., Arridge,S.R. (2002). Detection and modeling of non-Gaussian apparent diffusion coefficient profiles in human brain data. Magnetic Resonance in Medicine 48(2), 331-340. ISSN: 0740-3194
2.17 EPSRC EngD studentship (Sortex). 1/11/2004--31/10/2007.
- Personnel: Chris Senanayake, Daniel Alexander
- Summary: This project developed methods to identify colours of objects in CCD images independent of lighting conditions. It had applications in the food industry.
- Selected Publications:
- Senanayake,C., Alexander,D.C. (2007). Colour transfer by feature-based histogram registration. British Machine Vision Conference, 1, 429-438
2.18 EPSRC EngD studentship (Philips Medical Systems). 1/10/04--30/9/07.
- Personnel: Shahrum Nedjati-Gilani, Daniel Alexander
- Summary: This project aimed to develop techniques for identifying fibre crossing in human diffusion MRI scans. Shahrum's thesis shows experiments with model selection algorithms to distinguish one, two and three fibre crossings. He also develops a model-based super-resolution algorithm that can identify the interfaces between fibres to subvoxel accuracy.
- Selected Publications:
- Nedjati-Gilani, S., Alexander, D.C., (2009). Models for fanning and bending sub-voxel structures in diffusion MRI. Proceedings of the DMFC MICCAI 2009 Workshop. London, UK.
- Nedjati-Gilani, S. (2008). Reconstruction of Complex White Matter Architecture from Diffusion MRI. University of London PhD Thesis.
- Nedjati-Gilani,S., Parker,G.J., Alexander,D.C. (2008). Regularized Super-Resolution for Diffusion MRI. IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI) , 875-878
2.19 EPSRC Fast-stream grant. "Registration of DT MRI''. GR/R13715/01. 1/2/2001--31/1/2004.
- Personnel: Kathleen Curran, Daniel Alexander
- Summary: This project studied techniques for registration of diffusion tensor images. Registration of these images is complicated by the orientational information at each image voxel, which must be preserved through warps of the image. The TMI 2001 paper below provides algorithms for preserving orientation through image warps. Later papers describe how we can exploit that information for improving image matching and registration.
- Selected Publications:
- Zhang,H., Yushkevich,P.A., Alexander,D.C., Gee,J.C. (2006). Deformable registration of diffusion tensor MR images with explicit orientation optimization. Medical image analysis 10(5), 764-785. ISSN: 1361-8415
- Gee,J.C., Alexander,D.C. (2006). Diffusion tensor image registration. Chapter 20 in Weichert,J., Hagen,H. (ed.) Visualization and processing of tensor fields. Mathematics and Visualisation series. Springer, 327-344. ISBN: 978-3-540-25032-6
- Alexander,D.C., Pierpaoli,C., Basser,P.J., Gee,J.C. (2001). Spatial transformations of diffusion tensor magnetic resonance images. IEEE Transactions on Medical Imaging 20(11), 1131-1139. ISSN: 0278-0062
- Alexander,D.C., Gee,J.C., Bajcsy,R. (1999). Similarity measures for matching diffusion tensor images. British Machine Vision Conference (BMVC)
2.20 EPSRC/DTI E-science intiative. 1/10/2001--31/3/2002.
- Personnel: Phil Cook, Chris Parker, Anthony Steed, Daniel Alexander.
- Summary: This project built an interactive visualization for diffusion tensor imaging data in an immersive virtual environment. Users view and navigate around the data within a virtual reality CAVE and can collaborate with other users viewing the same data set at remote locations. The system has potential for education and remote consultation. It was demonstrated at the opening of the National E-science Centre in April 2002.
- Selected Publications:
- Steed,A., Alexander,D., Cook,P., Parker,C. (2003). Visualizing Diffusion-Weighted MRI Data Using Collaborative Virtual Environment and Grid Technologies. Theory and Practice of Computer Graphics, IEEE Computer Society

